GPU Computing in Julia¶
This session introduces GPU computing in Julia.
GPGPU¶
GPUs are ubiquitous in modern computers. Following are GPUs today's typical computer systems.
| NVIDIA GPUs | Tesla K80 | GTX 1080 | GT 650M |
|---|---|---|---|
 |
 |
 |
|
| Computers | servers, cluster | desktop | laptop |
 |
 |
 |
|
| Main usage | scientific computing | daily work, gaming | daily work |
| Memory | 24 GB | 8 GB | 1GB |
| Memory bandwidth | 480 GB/sec | 320 GB/sec | 80GB/sec |
| Number of cores | 4992 | 2560 | 384 |
| Processor clock | 562 MHz | 1.6 GHz | 0.9GHz |
| Peak DP performance | 2.91 TFLOPS | 257 GFLOPS | |
| Peak SP performance | 8.73 TFLOPS | 8228 GFLOPS | 691Gflops |
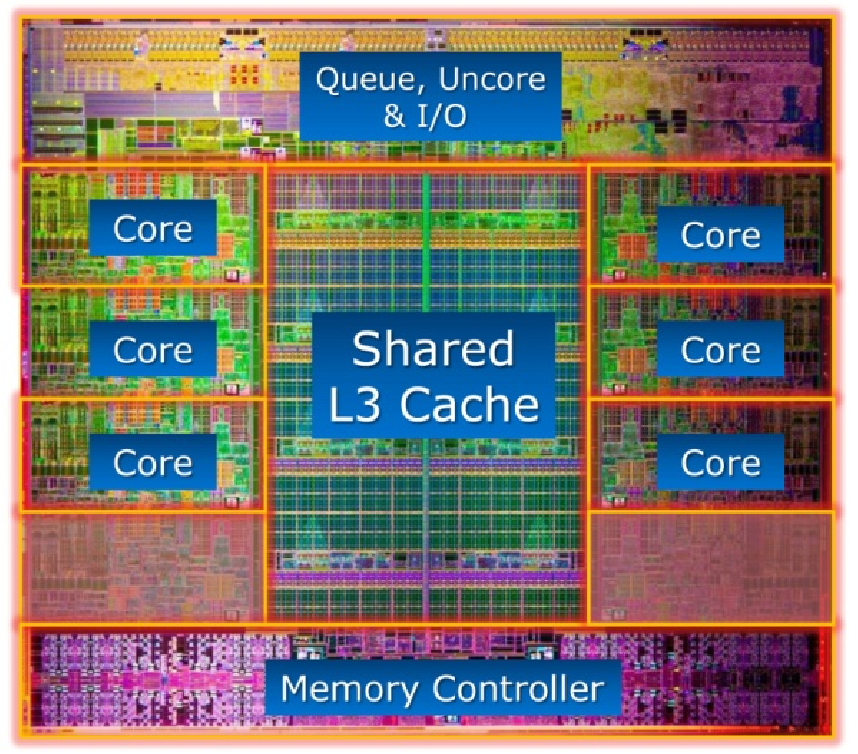
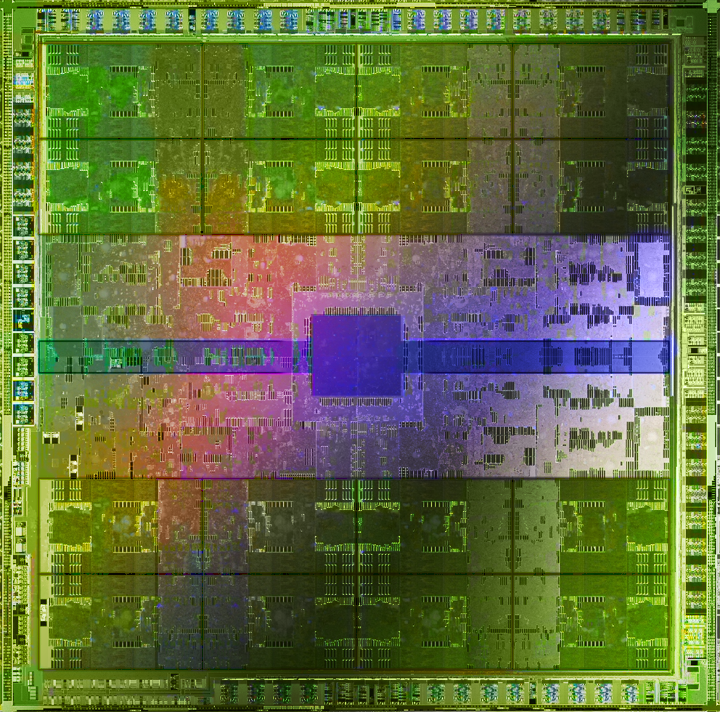
GPU architecture vs CPU architecture.
- GPUs contain 100s of processing cores on a single card; several cards can fit in a desktop PC
- Each core carries out the same operations in parallel on different input data -- single program, multiple data (SPMD) paradigm
- Extremely high arithmetic intensity if one can transfer the data onto and results off of the processors quickly
 |
 |
|---|---|
 |
 |
GPGPU in Julia¶
GPU support by Julia is under active development. Check JuliaGPU for currently available packages.
There are at least two paradigms to program GPU in Julia.
CUDA is an ecosystem exclusively for Nvidia GPUs. There are extensive CUDA libraries for scientific computing: CuBLAS, CuRAND, CuSparse, CuSolve, CuDNN, ...
The CUDA.jl package allows defining arrays on Nvidia GPUs and overloads many common operations.
The AMDGPU.jl package allows defining arrays on AMD GPUs and overloads many common operations.
Warning: Most recent Apple operating system iOS 10.15 (Catalina) does not support CUDA yet.
I'll illustrate using CuArrays on my Linux box running CentOS 7. It has a NVIDIA GeForce RTX 2080 Ti OC with 11GB GDDR6 (14 Gbps) and 4352 cores.
versioninfo()
Query GPU devices in the system¶
using CUDA
CUDA.versioninfo()
[CUDA.capability(dev) for dev in CUDA.devices()]
Transfer data between main memory and GPU¶
# generate data on CPU
x = rand(Float32, 3, 3)
# transfer data form CPU to GPU
xd = CuArray(x)
# generate array on GPU directly
yd = CUDA.ones(3, 3)
# collect data from GPU to CPU
x = collect(xd)
Linear algebra¶
using BenchmarkTools, LinearAlgebra
n = 1024
# on CPU
x = rand(Float32, n, n)
y = rand(Float32, n, n)
z = zeros(Float32, n, n)
# on GPU
xd = CuArray(x)
yd = CuArray(y)
zd = CuArray(z)
# SP matrix multiplication on GPU
@benchmark mul!($zd, $xd, $yd)
# SP matrix multiplication on CPU
@benchmark mul!($z, $x, $y)
We see ~40-50x fold speedup in this matrix multiplication example.
# cholesky on Gram matrix
xtxd = xd'xd + I
@benchmark cholesky($(Symmetric(xtxd)))
xtx = collect(xtxd)
@benchmark cholesky($(Symmetric(xtx)))
GPU speedup of Cholesky on this example is moderate.
Elementiwise operations on GPU¶
# elementwise function on GPU arrays
fill!(yd, 1)
@benchmark $zd .= log.($yd .+ sin.($xd))
# elementwise function on CPU arrays
x, y, z = collect(xd), collect(yd), collect(zd)
@benchmark $z .= log.($y .+ sin.($x))
GPU brings great speedup (>500x) to the massive evaluation of elementary math functions.